Wrestling with SUMO
Have you ever thought of using a solubility tag to enhance expression of your protein, considered using SUMO and then wondered, which sequence should I use? and which version is most suitable for the expression host I’m using? It’s certainly a question we’ve asked ourselves as there seem to be a different variations that people have described in publications as well as commercially available plasmids containing SUMO tags. Having considered what the different versions are, we thought we share our thoughts on this very useful solubility tag.
SUMO as a Solubilisation Tag
Small Ubiquitin-related Modifier (SUMO) proteins (Smt3 in yeast and SUMOs 1-4 in vertebrates) are 11.6kDa ubiquitin-like proteins (Ulps), that can post-translationally modify target proteins. The target proteins, of which there are thousands, have specific sumoylation motifs, and modification with SUMO can influence their solubility, stability, cellular localisation and interactions. This leads to SUMO having a key influence in cellular process such as cell proliferation, genomic stability and infection.
In a similar manner to ubiquitin, SUMO is covalently attached to lysine side chains on target proteins. The enzyme pathway that achieves this operates in a cycle (in Figure 1 below); the precursor SUMO is processed by SUMO proteases (e.g. Ulp1 in yeast), which remove the C-terminal residues to expose a conserved diglycine motif and subsequently the SUMO is transferred to the target protein via an E1 activating enzyme, E2 conjugating enzyme and E3 ligase.
While this process is biochemically similar to that for ubiquitin attachment to substrates targeted for proteolytic degradation, the SUMO proteases, E1, and E2 are specific for SUMO and do not function with ubiquitin or other Ulps.
Ultimately these sumoylated proteins are further processed by the cellular SUMO specific proteases, such as Ulp1, which cleave and deconjugate SUMO from the side chain lysines.
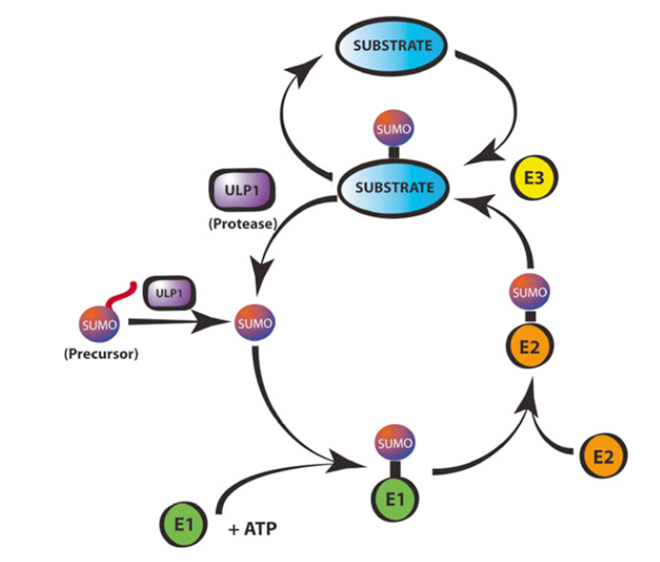
Figure 1: The sumoylation cycle (taken from [2])
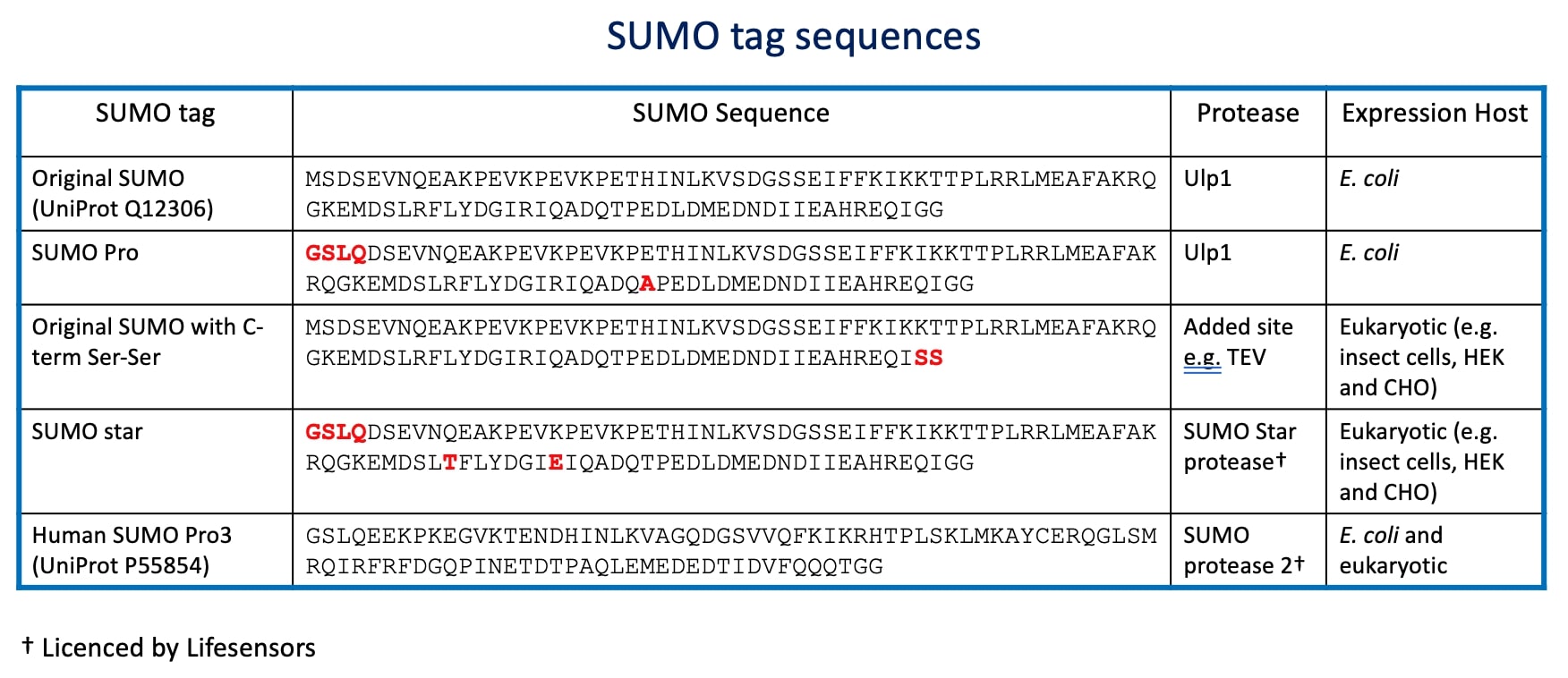
Malakhov et al 2004 [1] were the first to describe the attachment of SUMO to the N-terminus of a protein sequence to improve soluble expression yields of target proteins expressed in E.coli. The particular version used was the mature form of Smt3 from yeast (Uniprot reference Q12306). Lifesensors also have an alternative version they call SUMOPro which has a T77A mutation and can also be used for expression in E.coli.
What’s So Good About the SUMO Tag and Why Do We Use It?
Well, firstly, from our experience, it does indeed improve soluble expression in E.coli and more importantly, the target protein often remains soluble post tag cleavage, suggesting that SUMO facilitates the initiation of translation and has chaperone-like functions [1], [2]. Given SUMO’s relatively small size, it can also be used with other affinity tags such as 6His tor Twin Strep to facilitate purification.
Another key advantage is that the protease that removes the SUMO tag, Ulp1, is very specific and efficient (~25x better Kcat than TEV protease [3]). Cleavage is after the SUMO C-terminal Gly-Gly residues and therefore leaves no carry over residues on the target. Most other common protease sites used in recombinant proteins do leave residues which for some applications, such as crystallography, could be undesirable (see our Protease Information Protocol for details).
As a cleavage protease, Ulp1 has the advantage that it is easy to produce recombinantly. This is in contrast to the de-ubiquitinating proteases of other members of the small ubiquitin-like family, which are often unstable and difficult to produce recombinantly and consequently are not available commercially.
Inevitably, no tag is perfect and the downsides of SUMO tags are that generally it is only useful as an N-terminal fusion; the P1’ position can’t be a Proline (Lys, Val and Leu are also undesirable) and eukaryotic expression hosts (such as insect cells, HEK and CHO cells) contain native de-SUMOlyases which can remove recombinant SUMO tags, reducing the yields obtained.
However, this latter issue has been overcome with the development of the SUMOstar tag [4] which is a R64T,R71E double mutant version of the Yeast Smt3 SUMO that is resistant to eukaryotic de-SUMOylases. Unfortunately, it was also resistant to Ulp1, so the authors developed a mutant Ulp1, SUMOstar protease, that is capable of cleaving SUMOstar. These are now commercially available from Lifesensors who also produce other SUMO tag options including one based on human SUMO3 (see table below).
Another way to overcome the native de-SUMOlyase issue in eukaryotic hosts is to just mutate the C-terminal Gly-Gly residues to Ser-Ser. This means that the Ulp1 can’t cleave the tag, so another protease site (e.g. TEV) has to be added on. However, the SUMO tag can still function to improve soluble expression yields with many target recombinant protein constructs and is therefore still a useful tool to consider when designing constructs.
For further information on other commonly used protein tags we use at Peak Proteins, please have a read of our our Protein Tag and Leader Sequence Protocol . To find out more about our protein expression and purification services do get in contact with us and we’d be happy tell you more.
References
[1] M. P. Malakhov, M. R. Mattern, O. A. Malakhova, M. Drinker, S. D. Weeks, and T. R. Butt, ‘SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins’, J. Struct. Funct. Genomics, vol. 5, no. 1/2, pp. 75–86, Mar. 2004, doi: 10.1023/B:JSFG.0000029237.70316.52.
[2] D. Kuo, M. Nie, and A. J. Courey, ‘SUMO as a Solubility Tag and In Vivo Cleavage of SUMO Fusion Proteins with Ulp1’, 2014, pp. 71–80.
[3] J. G. Marblestone, ‘Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO’, Protein Sci., vol. 15, no. 1, pp. 182–189, Jan. 2006, doi: 10.1110/ps.051812706.
[4] L. Liu, J. Spurrier, T. R. Butt, and J. E. Strickler, ‘Enhanced protein expression in the baculovirus/insect cell system using engineered SUMO fusions’, Protein Expr. Purif., vol. 62, no. 1, pp. 21–28, Nov. 2008, doi: 10.1016/j.pep.2008.07.010.
[5] A. B. Celen and U. Sahin, ‘Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts’, FEBS J., vol. 287, no. 15, pp. 3110–3140, Aug. 2020, doi: 10.1111/febs.15319.
[6] V. G. Wilson and P. R. Heaton, ‘Ubiquitin proteolytic system: focus on SUMO’, Expert Rev. Proteomics, vol. 5, no. 1, pp. 121–135, Feb. 2008, doi: 10.1586/14789450.5.1.121.


